Understanding marine mammal distributions and habitat use through environmental DNA

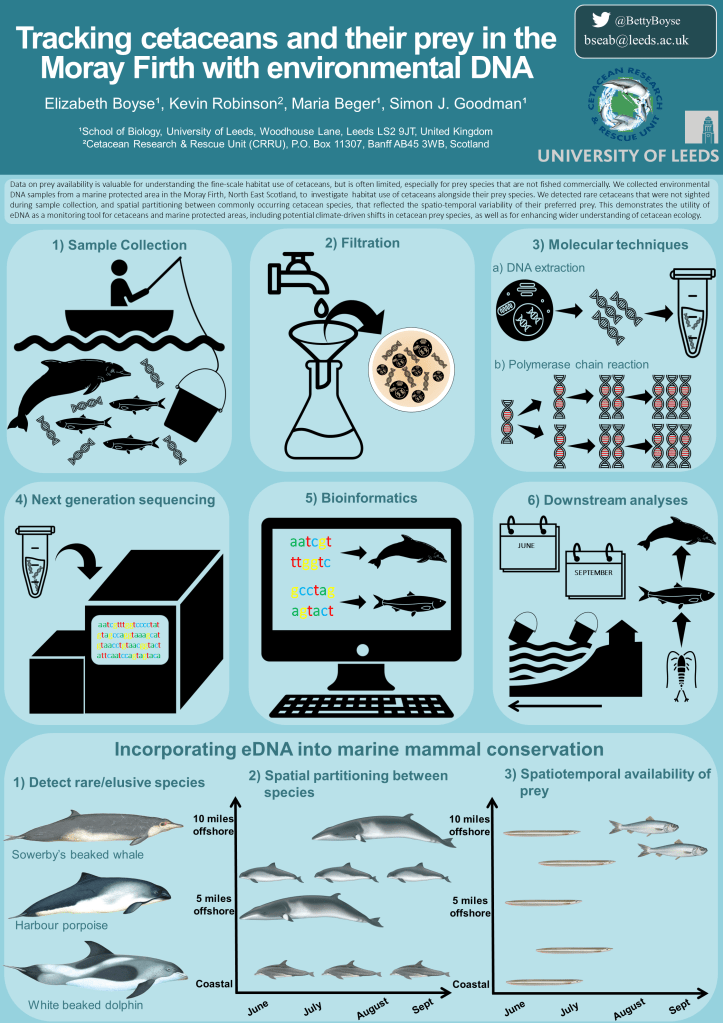
Environmental DNA (eDNA) is developing rapidly as an important tool for biodiversity monitoring and for exploring ecological interactions. eDNA is especially useful for uncovering species presence and distributions for elusive or hard to observe species in remote locations, or environments that are logistically challenging to access. We have recently demonstrated the power of eDNA for monitoring of common and rare cetaceans (including Minke whales, harbour porpoise, bottlenosed dolphins and Sowerby’s beaked whale) in a marine protected area in the Moray Firth, northeast Scotland, UK, and for uncovering spatio-temporal patterns in species presence and ecological interactions with their prey.
We’re now aiming to build on our current work to expand the spatial and temporal coverage for the Moray Firth and other areas with the aims of:
- Evaluating the environmental and ecological factors influencing spatial and temporal habitat use by minke whales, harbour porpoise and bottlenose dolphins
- Explore the drivers of spatio-temporal variation in marine community structure and species interactions
- Develop and test improved methods for marine eDNA sampling to expand spatio-temporal coverage
- Develop and improve methods for reconstructing ecological interactions using eDNA (e.g. food webs and co-occurrence analyses)
The project will be co-supervised by Dr Maria Beger and Elizabeth Boyse (University of Leeds), Dr Kevin Robinson (Cetacean Research and Rescue Unit), Dr Elena Valsecchi (University of Milano Biocca), and potentially other collaborators from the UK cetacean eDNA network which is currently under development.
PhD funding & eligibility
For PhDs, a variety of competitive scholarships are available which support UK home rated students (UK citizens, and those satisfying relevant UK residency criteria), for example the Leeds Doctoral Scholarship. Alternatively we can consider overseas students supported by external scholarships, or self-funded home and overseas students.
How to apply: Applications will be made via the Faculty of Biological Sciences Graduate School Portal (https://biologicalsciences.leeds.ac.uk/research-degrees), but it is essential that you email me (s.j.goodman@leeds.ac.uk) before submitting an application.
Application deadlines: Various, from 7th January 2026
Post-doctoral fellowships
I am also happy to hear from post-doctoral candidates seeking to develop independent fellowships (e.g. Marie-Curie fellowships) to work in this research area at the University of Leeds. Please contact me to discuss proposal ideas and potential funding opportunities.
Related publications
- Boyse, E., Robinson, K.P., Carr, I.M., Valsecchi, E., Beger, M. and Goodman, S.J. (2025), Inferring Species Interactions From Co-occurrence Networks With Environmental DNA Metabarcoding Data in a Coastal Marine Food Web. Mol Ecol, 34: e17701. https://doi.org/10.1111/mec.17701
- Boyse E, Robinson KP, Beger M, Carr IM, Taylor M, Valsecchi E, Goodman SJ (2024). Environmental DNA reveals fine-scale spatial and temporal variation of marine mammals and their prey species in a Scottish marine protected area. Environmental DNA 6, e587. https://doi.org/10.1002/edn3.587.
- Boyse, E., Beger, M., Valsecchi, E., & Goodman, S. J. (2023). Sampling from commercial vessel routes can capture marine biodiversity distributions effectively. Ecology and Evolution, 13, e9810. https://doi.org/10.1002/ece3.9810
- Valsecchi E, Arcangeli A, Lombardi R, Boyse E, Carr IM, Galli P, Goodman SJ. 2021. Ferries and Environmental DNA: Underway Sampling From Commercial Vessels Provides New Opportunities for Systematic Genetic Surveys of Marine Biodiversity. Frontiers in Marine Science. 8
- Valsecchi E, Bylemans J, Goodman SJ, Lombardi R, Carr I, Castellano L, Galimberti A, Galli P. 2020. Novel universal primers for metabarcoding environmental DNA surveys of marine mammals and other marine vertebrates. Environmental DNA. 2(4), pp. 460-476